FLASHQuant performs MS1-level label-free quantification data analysis in top-down proteomics with an automatic overlapping signal resolution method.
Input/Output
- Input: Centroided MS1 scans (*.mzML)
- Output: Quantified proteoforms in a tab-separated file (*.mzML); optionally, OpenMS LC-MS features output (*.featureXML)
- for each proteoform, mono-isotopic/average mass, retention time range, charge range, different types for quantity values, and isotope cosine similarity score are provided.
- Parameters can be found by running FLASHQuant using the “–helphelp” option.
Consensus FeatureGroup (putative proteoform) detection
FLASHQuant runs per scan; thus, we provide an additional simple tool for detecting consensus feature groups among multiple scans (i.e., technical replicates) named TopDownConsensusFeatureGroup. Retention time and mass tolerance can be adjusted with parameters.
GUI
FLASHQuant and (consecutive) TopDownConsensusFeatureGroup can be executed easily with GUI, FLASHQuantWizard.
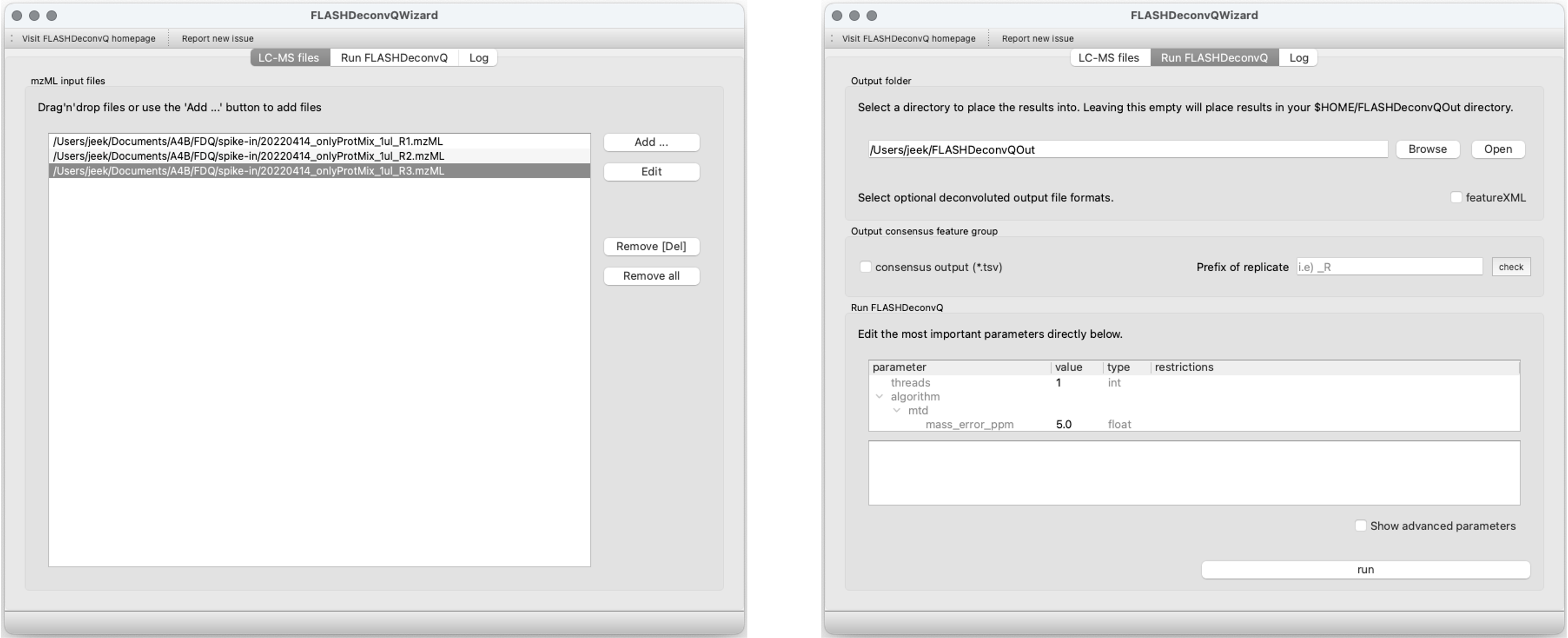
To learn how to run the programs, please check FLASHDeconv page for details.